ParSAC calibration slowing down when printing output to console
September 11, 2020
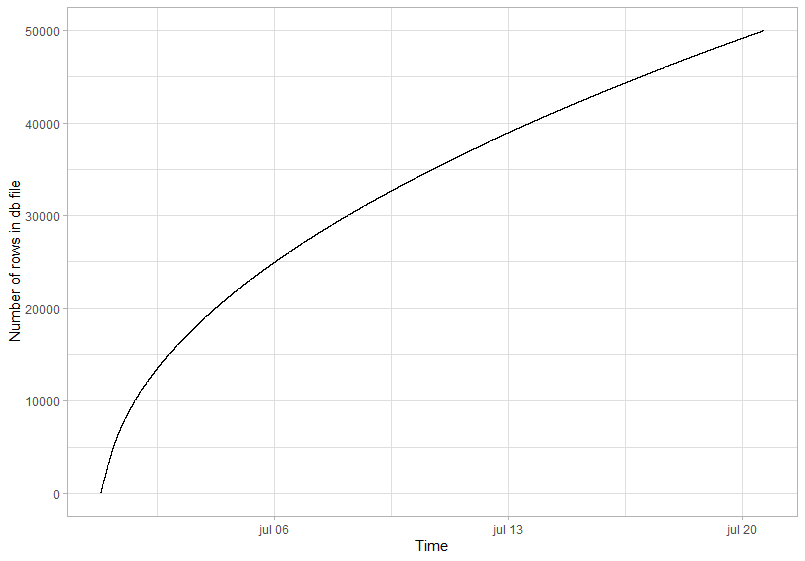
I recently noticed that the ParSAC calibration of my coupled GOTM-FABM-biogeochemical model was slowing down over time (see first figure). Since I tend to use 150,000+ iterations, calibrations take a long while (i.e. more than 1 day), but I didn’t understand why it slowed down. Karsten and Jorn pointed out that a memory leak might be the potential cause.
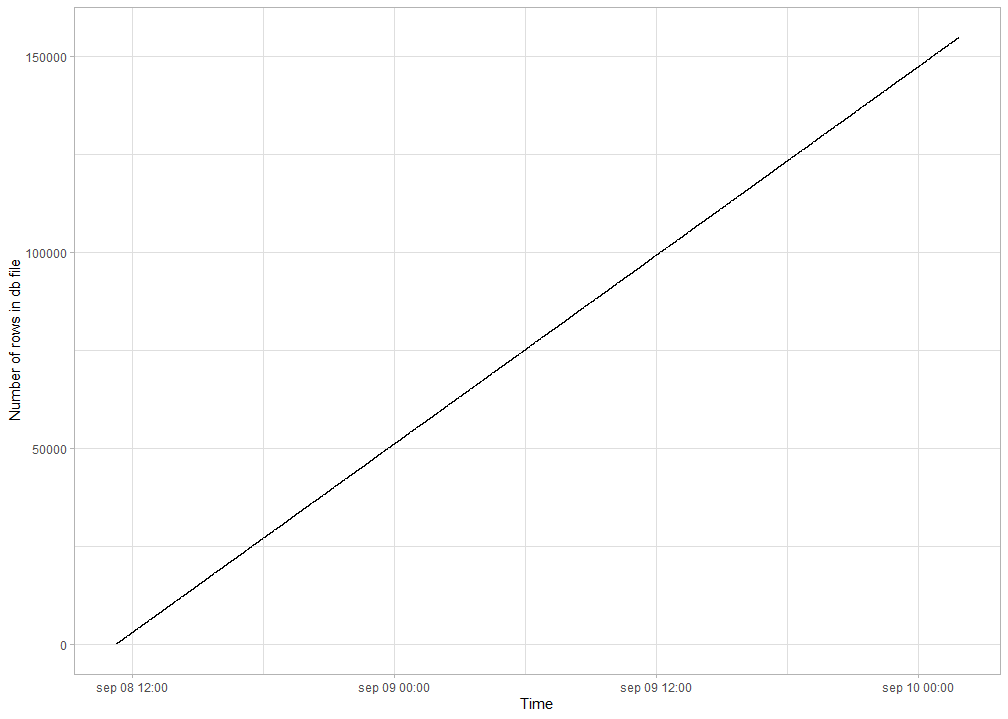
ParSAC was called through the system-function in the R software and I had set the show.output.on.console arguement to TRUE. This printed the text output of the ParSAC calibration to the Rstudio console, but caused increasing memory use over time. By simply setting the argument to FALSE, the calibration no longer slowed down over time (see second figure), and speed markedly improved (for a test case with the same number of calibrations, a speed up of 157 hours to 44 hours).
The aim of this short blog post is to make users aware that printing output to console is a potential bottleneck for the speed of the ParSAC calibration. An indication of a memory leak is the number of entries in your database file increasing slower and slower over time. Fortunately, the solution is relatively straightforward once you figure out what is the issue.